The BraiNum project aims at developing a mathematical model for the physiological and pathological function of the brain and central nervous system. In this endeavor, we exploit the power and flexibility of mathematical modeling, numerical analysis, and high-performance scientific computing.
BrainNum project is developed at the Laboratory for Modeling and Scientific Computing (MOX) of the Department of Mathematics, Politecnico di Milano.
We focus on:
- Neurodegenerative diseases: high incidence (about 1/1000 subjects worldwide) and have a major clinical and societal impact on national systems. Their investigation and treatment are still the subject of open discussion in the medical community, and their study is ever more relevant, due to the overall aging of the world population.
- Neurological disorders: broad spectrum of pathological conditions related to the impairment of neural firing rate or signal transmission. Epilepsy (the most common) affects 50 million people worldwide, and acute seizures entail possibly severe physical injuries.
Research Team

Paola F. Antonietti
(Principal Investigator)

Francesco Daniele

Beatrice Caon
Mathematical modeling of the brain and CNS
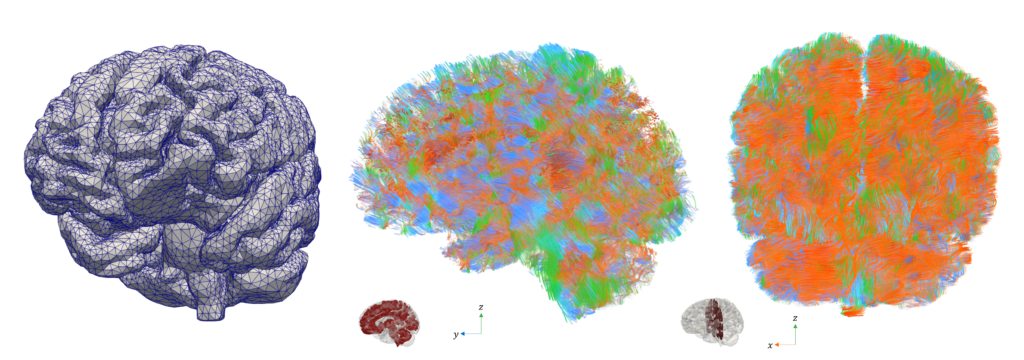
The computational modeling of the Central Nervous System (CNS) entails several challenges:
- anatomic complexity: cerebral gyri and sulci, ventricles, vasculature, and ducti
- multiphysics: electrophysiology, blood supply and perfusion, dynamics of the cerebrospinal fluid (CSF), protein distribution and clearance, long-term remodeling
- multiscale: from single cell (neuron firing, axon mechanical remodeling, …) to the whole organ (signal propagation, blood and CSF dynamics, cerebral tissue poroelasticity, prion clearance, …); from milliseconds (electrical activation) to decades (neurodegeneration and remodeling)
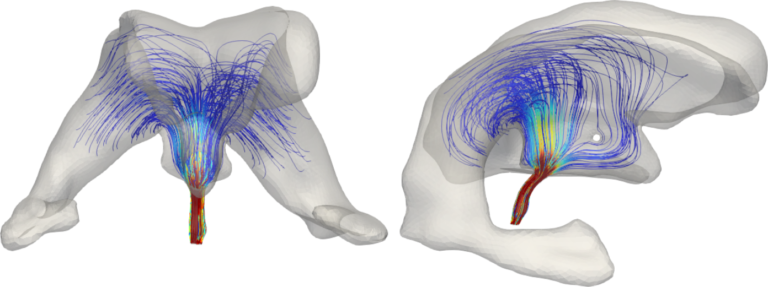
![MPETCompartimenti Arterial pressure, CSF pressure, and tissue displacement resulting from a multiple network poroelasticity simulation in physiological conditions [Corti, Antonietti, Dede’, Quarteroni. M3AS, accepted (2023)]<br />](https://brainum.mox.polimi.it/wp-content/uploads/2023/05/MPETCompartimenti.png)
Arterial pressure, CSF pressure, and tissue displacement resulting from a multiple network poroelasticity simulation in physiological conditions [Corti, Antonietti, Dede’, Quarteroni. M3AS, accepted (2023)]
![MPETCompartimenti Arterial pressure, CSF pressure, and tissue displacement resulting from a multiple network poroelasticity simulation in physiological conditions [Corti, Antonietti, Dede’, Quarteroni. M3AS, accepted (2023)]<br />](https://www.mate.polimi.it/servizi/biblioteca/quaderni/upload/img/mox-img-20231011185231.png)
Intracranial pressure and parenchymal displacement obtained simulating the coupling between the CSF in the brain ventricles and the surrounding poroelastic tissue [Fumagalli, Corti, Parolini, Antonietti. arXiv:2310.07651 (2023)]
Neurodegenerative diseases
Modeling of:
- CSF flow: waste clearance and its impairment in neurodegeneration (time scale: seconds/minutes)
- distribution of misfolded proteins (time scale: years)
- neural axon degeneration and brain remodeling (time scale: decades)
- anatomic complexity: cerebral gyri and sulci, ventricles, vasculature, and ducti
- multiphysics: electrophysiology, blood supply and perfusion, dynamics of the cerebrospinal fluid (CSF), protein distribution and clearance, long-term remodeling
- multiscale: from single cell (neuron firing, axon mechanical remodeling, …) to the whole organ (signal propagation, blood and CSF dynamics, cerebral tissue poroelasticity, prion clearance, …); from milliseconds (electrical activation) to decades (neurodegeneration and remodeling)
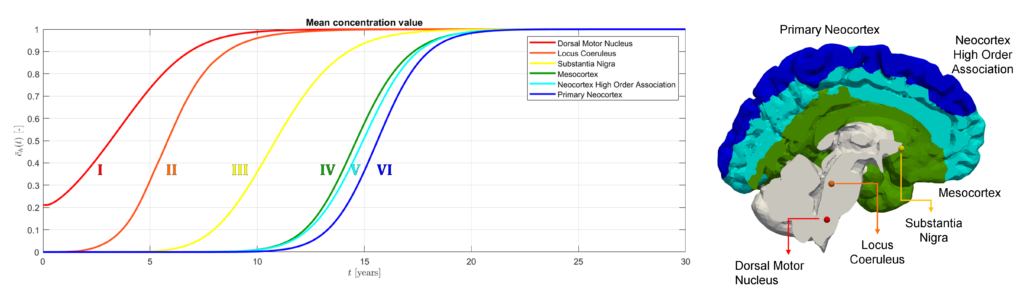
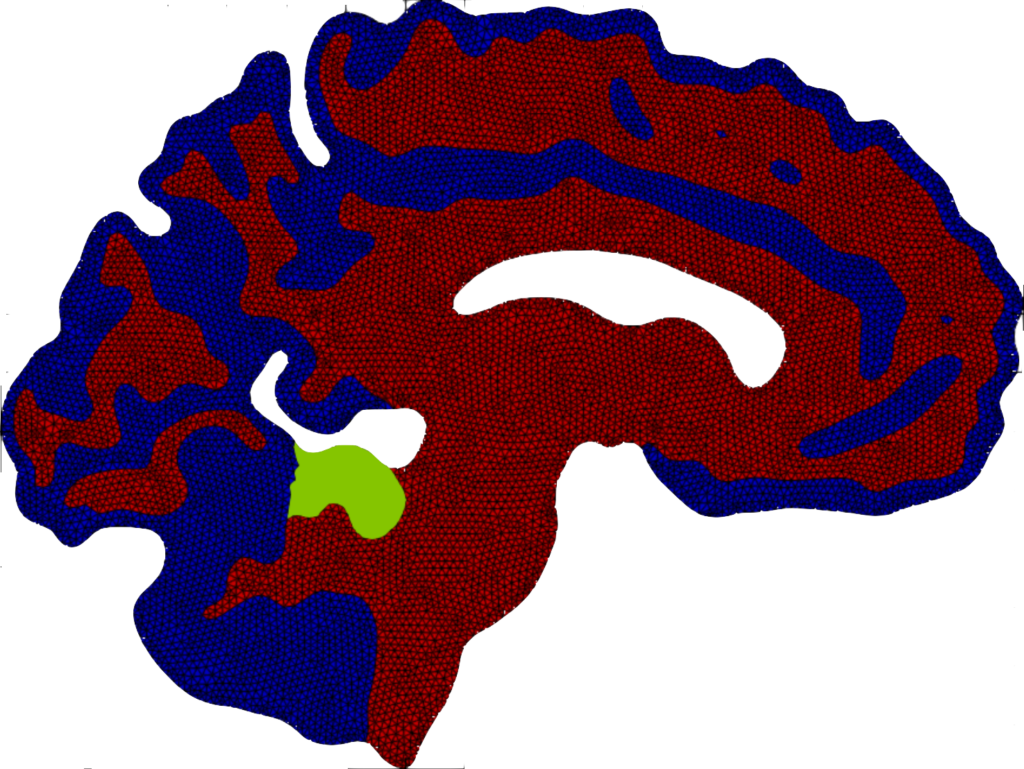
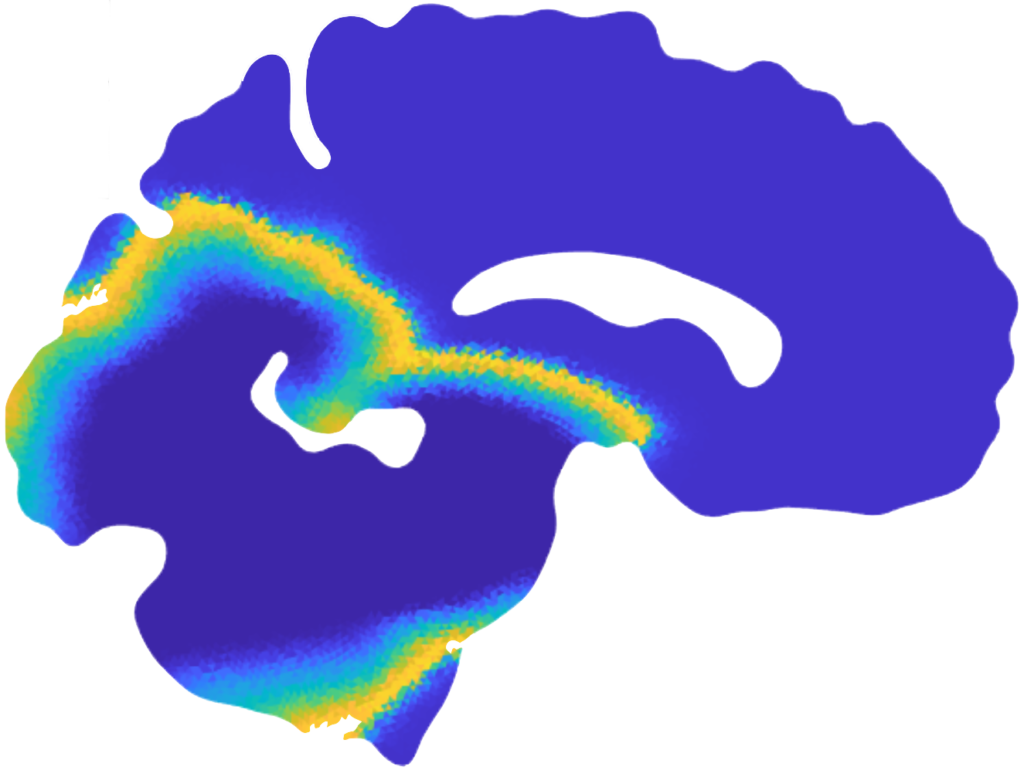
Mean value of the prions’ concentration inside some selected regions of the brain in Parkinson’s disease over 30 years (left) and position of brain regions (right) [Corti et al. (2023)].
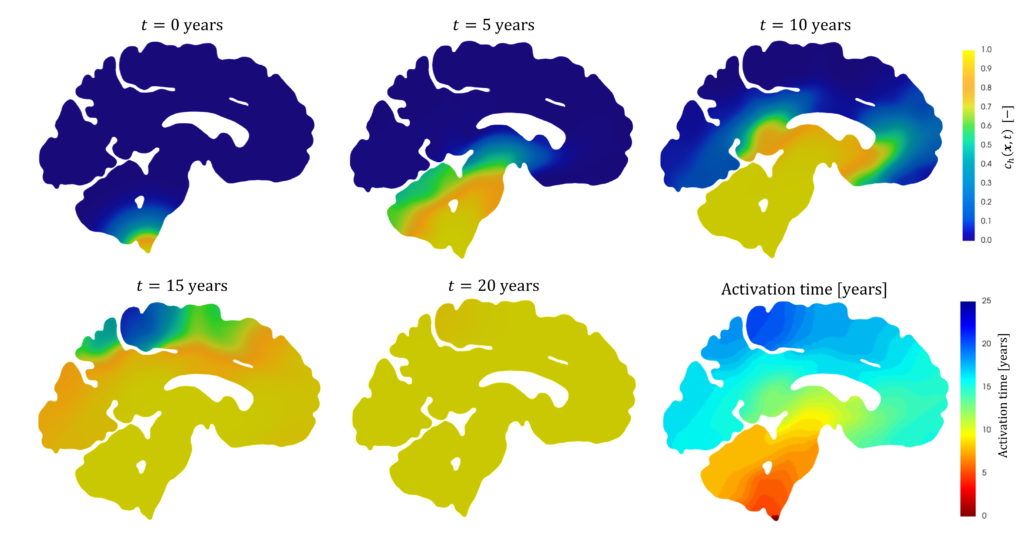
Patterns of α-synuclein concentration in Parkinson’s disease at different stages of the pathology and activation time of the pathology (bottom-right) [Corti et al. (2023)].
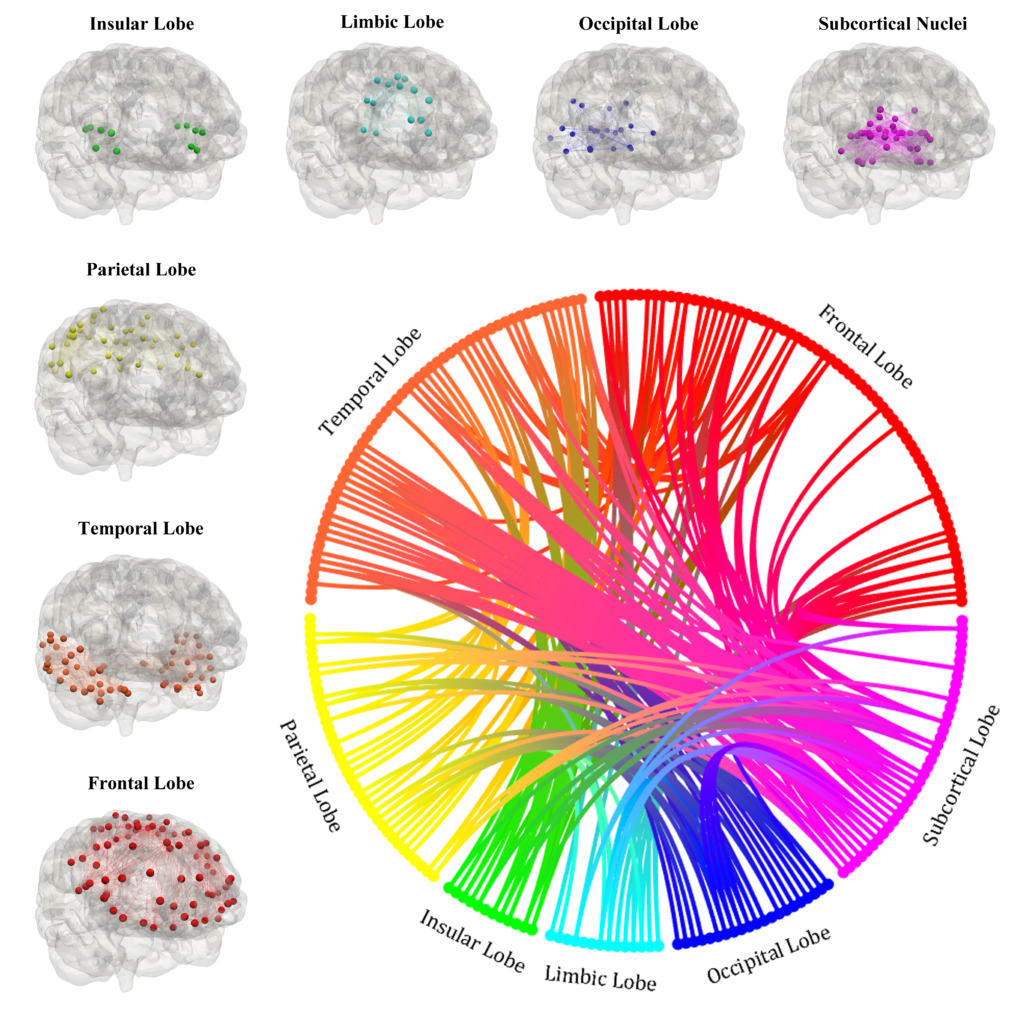
Local graphs of the seven regions of the brain and brain connectogram between different regions [Corti et al. (2023)].
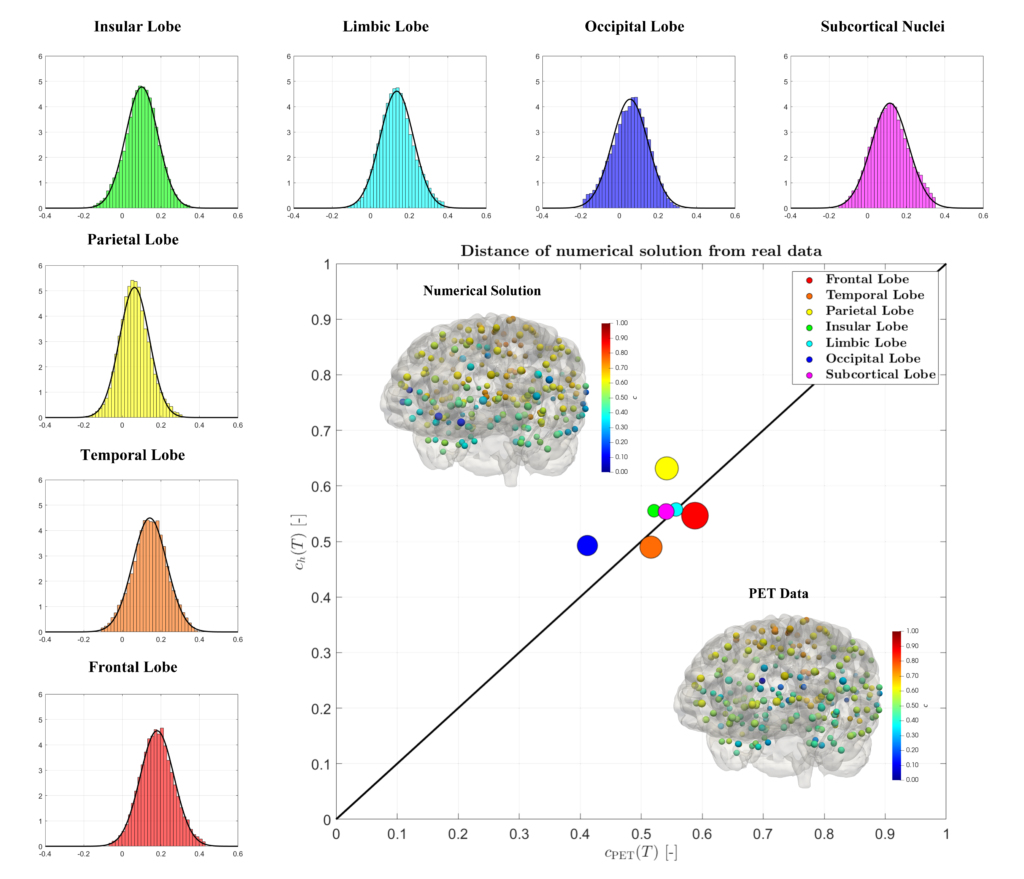
Histogram and Gaussian distribution associated with the reaction parameters of amyloid-β, in each lobe of the brain and comparison between the medical data and the numerical results for the concentration on a connectome [Corti et al. (2023)].
Neurological disorders
Numerical modelling of epileptic seizures. Epilepsy is a clinical neurological disorder characterized by recurrent and spontaneous seizures generating an abnormal high-frequency electrical activity of the brain. Despite several clinical studies that have permitted the development of specific pharmacological and surgical treatments to control the onset of seizures there is still an open debate on the mechanisms and optimal patient-specific treatment. We consider the bidomain model to mathematically describe seizure evolution in the grey and white matter, coupled with specific ionic models for neuronal modelling in which the different dynamics of the potential and ionic currents are considered. The mathematical model is discretized by means of discontinuous Galerkin methods. Suitable (space-time) adaptive schemes are proposed to exploit the flexibility of the numerical approach to better describe the sharp propagating fronts that characterize seizure propagation. The numerical results provide insights into the mechanisms underlying seizures and might, in the future, support precision medicine, thanks to methods capable of predicting patient-specific behaviour and suggesting optimal treatment.

Publications
Polytopal mesh agglomeration via geometrical deep learning for three-dimensional heterogeneous domains
Authors: P. F. Antonietti, M. Corti, G. Martinelli
Journal Reference: Mathematics and Computers in Simulation, 241(part B). 335-353 (2026)
A structure-preserving LDG discretization of Fisher-Kolmogorov equation for modeling neurodegenerative diseases
Authors: P.F. Antonietti, M. Corti, S. Gómez, I. Perugia
Journal reference: Mathematics and Computers in Simulation. 241(part A). 351-366 (2026)
A p-adaptive polytopal discontinuous Galerkin method for high-order approximation of brain electrophysiology
Authors: Caterina B. Leimer Saglio, Stefano Pagani, P.F. Antonietti
Journal reference: Computer Methods in Applied Mechanics and Engineering. 446(part A). 118249 (2025)
A high-order discontinuous Galerkin method for the numerical modeling of epileptic seizures
Authors: C. B. Leimer Saglio, S. Pagani, M. Corti, P. F. Antonietti
Journal Reference: Computers & Mathematics with Applications, 205. 112-131 (2026)
Numerical modelling of protein misfolding in neurodegenerative diseases: a computational study
Authors: P.F. Antonietti, M. Corti
Chapter reference: Numerical Mathematics and Advanced Applications ENUMATH 2023, Volume 1. Springer (2025)
Discontinuous Galerkin method for a three-dimensional coupled fluid-poroelastic model with applications to brain fluid mechanics
Authors: I. Fumagalli
Journal reference: Mathematics in Engineering. 7 (2). 130-161 (2025)
A posteriori error analysis for a coupled Stokes-poroelastic system with multiple compartments
Authors: I. Fumagalli, N. Parolini, M. Verani
Journal reference: Journal of Scientific Computing. 103. 22 (2025)
lymph: discontinuous poLYtopal methods for Multi-PHysics differential problems
Authors: P.F. Antonietti, S. Bonetti, M. Botti, M. Corti, I. Fumagalli, I. Mazzieri
Journal reference: ACM Transactions on Mathematical Software. 51 (1). 3:1-3:22 (2025)
A coupled mathematical and numerical model for protein spreading and tissue atrophy applied to Alzheimer's disease
Authors: V. Pederzoli, M. Corti, D. Riccobelli, P. F. Antonietti
Journal reference: Computer Methods in Applied Mechanics and Engineering, 444. 118118 (2025)
Discontinuous Galerkin approximations of the heterodimer model for protein-protein interaction
Authors: P.F. Antonietti, F. Bonizzoni, M. Corti, A. Dall’Olio
Journal reference: Computer Methods in Applied Mechanics and Engineering. 431. 117282 (2024)
Exploring tau protein and amyloid-beta propagation: A sensitivity analysis of mathematical models based on biological data
Authors: M. Corti
Journal reference: Brain Multiphysics 7:100098 (2024)
Structure preserving polytopal discontinuous Galerkin methods for the numerical modeling of neurodegenerative diseases
Authors: M. Corti, F. Bonizzoni, P.F. Antonietti
Journal reference: Journal of Scientific Computing. 100. 39 (2024)
Uncertainty quantification for Fisher-Kolmogorov equation on graphs with application to patient-specific Alzheimer's disease
Authors: M. Corti, F. Bonizzoni, P.F. Antonietti, A.M. Quarteroni
Journal reference: ESAIM: Mathematical Modelling and Numerical Analysis. 58 (6). 2135-2154 (2024)
Discontinuous Galerkin methods for Fisher–Kolmogorov equation with application to α-synuclein spreading in Parkinson’s disease
Authors: M. Corti, F. Bonizzoni, L. Dede’, A.M. Quarteroni, P.F. Antonietti
Journal reference: Computer Methods in Applied Mechanics and Engineering. 417. 116450 (2023)
Numerical modeling of the brain poromechanics by high-order discontinuous Galerkin methods
Authors: M. Corti, P.F. Antonietti, L. Dede’, A.M. Quarteroni
Journal reference: Mathematical Models and Methods in Applied Sciences. 33(08). 1577-1609 (2023)
Polytopal discontinuous Galerkin discretization of brain multiphysics flow dynamics
Authors: I. Fumagalli, M. Corti, N. Parolini, P.F. Antonietti
Journal reference: Journal of Computational Physics. 513. 113115 (2024)
Preprints
A whole-brain model of amyloid beta accumulation and cerebral hypoperfusion in Alzheimer's disease
Authors: M. Corti, A. Ahern, A. Goriely, E. Kuhl, P. F. Antonietti
Preprint Reference: ArXiv preprint 2601.08478 (2026)
A massively parallel non-overlapping Schwarz preconditioner for PolyDG methods in brain electrophysiology
Authors: C. B. Leimer Saglio, S. Pagani, P. F. Antonietti
Preprint Reference: ArXiv preprint 2512.19536 (2025)
Mathematical and numerical modeling of coupled oxygen dynamics and neuronal electrophysiology
Authors: F. Daniele, C. B. Leimer Saglio, S. Pagani, P. F. Antonietti
Preprint Reference: Arxiv preprint 2509.22863 (2025)
Structure-preserving local discontinuous Galerkin discretization of conformational conversion systems
Authors: P. F. Antonietti, M. Corti, S. Gómez, I. Perugia
Preprint Reference: Arxiv preprint 2511.04830 (2025)
A novel mathematical and computational framework of amyloid-beta triggered seizure dynamics in Alzheimer's disease
Authors: C. B. Leimer Saglio, M. Corti, S. Pagani, P. F. Antonietti
Preprint Reference: ArXiv preprint 2511.10369 (2025)
Predicting Alzheimer's disease progression from sparse multimodal data by NeuralODE models
Authors: A. Zanin, S. Pagani, M. Corti, V. Crepaldi, G. Di Fede, P.F. Antonietti, and for the Alzheimer’s Disease Neuroimaging Initiative (ADNI)
Preprint Reference: Biorxiv preprint 2025.08.26.672412 (2025)
A discontinuous Galerkin method for the three-dimensional heterodimer model with application to prion-like proteins’ dynamics
Authors: P.F. Antonietti, M. Corti, G. Lorenzon
Preprint reference: ArXiv preprint 2407.16065 (2024)
PhD Theses
Mathematical Models and Numerical Methods for Neurodegenerative Diseases
Author: M. Corti
Advisor: P. F. Antonietti
PhD in Mathematical Models and Methods in Engineering, Politecnico di Milano
Year: 2025
Advanced computational methods for the numerical modeling of epileptic seizures
Author: C. B. Leimer Saglio
Advisors: P. F. Antonietti, S. Pagani
PhD in Mathematical Models and Methods in Engineering, Politecnico di Milano
Year: in progress
This project is partially supported by




– Italian Research Center on High Performance Computing, Big Data and Quantum Computing (ICSC), under the NextGenerationEU project.
– National Recovery and Resilience Plan (NRRP), Mission 4, Component 1 – Investment 3.4 and Investment 4.1 funded by the European Union.
– PRIN 2020, research grant n. 20204LN5N5 “Advanced polyhedral discretisations of heterogeneous PDEs for multiphysics problems” funded by MUR (National Coordinator).
– ERC Sinergy Grant NEMESIS “NEw generation MEthods for Numerical SimulationS”, project number 101115663, funded by European Research Council






